You’ve got two copies of each chromosome, having received one copy of each chromosome from your mother and one chromosome from your father (this is true for your autosomes, but not for your X, Y, and mitochondria). When it comes time to pass on your DNA to the next generation, you in turn package up a single copy of each chromosome into a sperm/egg. Sometimes you pass on either mum or dad’s copy of a chromosome at random, often though you pass on a mosaic consisting of the two chromosomes (a recombinant chromosome).

The question came up (via a Slate article) of what is the probability that by chance your parent entirely failed to pass any autosomal DNA from a grandparent to you (e.g. your father fails to pass on any autosomal genome from your paternal grandfather)? There are 22 autosomes, so if there was no recombination that would happen with probability 2 x 0.5^22=4.7×10^(-7). But this probability is very much lower with recombination, as a recombinant chromosome necessarily has material from both parents. A discussion of how to do this calculation with recombination came up via Mike Eisen on twitter [1].
In order for you to receive your parent to transmit the entire autosome only from one grandparent, your parent also have to transmit all of their chromosomes without recombination [2]. Recombination also makes this probability differs between the sexes. This is because the probability that a chromosome is transmitted without recombination depends on the sex of the individual, females recombine more than males and so are less likely to transmit a chromosome without recombination. The probability of a chromosome being transmitted without recombination also depends on the size of the chromosome, big chromosomes recombine more. For example, chromosome 1 has a 2% chance of being transmitted to the next generation by females, but a 7% chance of this happening in males. While chromosome 22, a much smaller chromosome, has a 37% chance of being transmitted with out recombination in females, and has a 44% chance in males (you can look up this frequencies in the supplement of a paper I wrote with Adi Fledel-Alon and other folks from Molly Przeworski’s lab).
To work out the probability of all chromosomes failing to be transmitted with recombination for a particular sex we simply multiple together the probability of each chromosome being transmitted without recombination [3]. Doing this, we find that the probability that a male transmits every chromosome without recombination is 8.8 x 10^(-16), and this probability is substantially lower in females at 2.8×10^(-23).
Then having not recombined on any chromosome that parent would have to also transmit every chromosome without recombination (with probability 4.7×10^(-7)). So the probability that your mother fails entirely to transmit any autosomal genetic material from a particular grandparent to you is 1.3×10^(-29), and your father does this with probability 4.2×10^(-22). So it’s pretty bloody unlikely.
Perhaps a more interesting question what is the distribution of the fraction of the autosomal genome that your parent transmits to you from a particular grandparent (e.g. your maternal grandmother)?
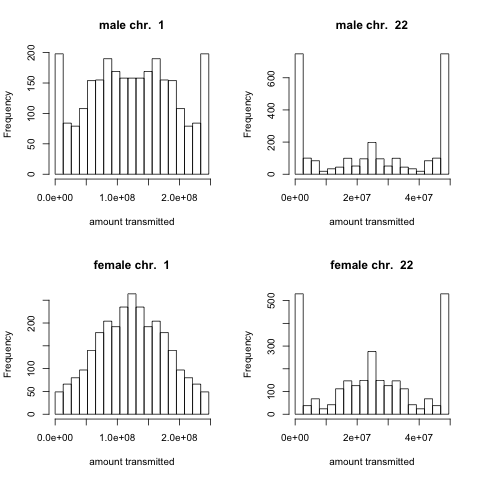
This question has been considered mathematically by a number of authors, as it has important applications for identifying unknown genetic relationships between individuals and estimating various heritability measures. However, to my knowledge no one has actually done this calculation using real recombination data (so I thought it would be fun to do). For each chromosome in turn, using recombination data from real transmissions, I simulated the amount of grandparental chromosome that was transmitted by a parent. For example, here’s the histogram of the distribution of the amount of chromosome 1 and 22 a father or a mother transmits.
These distributions are less variable in females than in males due to the greater number of recombination event in females than in males, and the fraction transmitted is more variable for small chromosomes as they have fewer recombination events. The pdf showing these histograms for every chromosome is here.
I then looked at what fraction of the entire (autosomal) genome from a particular grandparent was transmitted to the next generation.
I was a little surprised by how long tailed this was in males. Roughly 5/1000 fathers transmit less than 20% of one paternal grandparent’s autosome to the next generation!
Sometime soon I’ll generate these numbers for longer transmission chains, e.g. what’s the distribution of the fraction of your genome could you expect to receive from a great-grandparent.
1. I originally messed up this calculation, Mike Eisen got the right answer and pointed out my error. Thanks also to Amy Williams and Adam Auton for motivating some of the questions addressed here.
2. The probability of failing to transmit the entirety of one grandparental autosome is actually a lot lower than this, as gene conversion also can lead to transmission of small chunks of genome even if there is no crossing over. Gene conversion is thought to be ~10x as common as crossing over, and I estimate the probability of no transmitted crossovers or gene conversions to be <10^(-90). However, gene conversions are very small, so we might think the calculation above is for the bulk of the genome.
3. This isn’t quite right, as the recombination rates of different chromosomes aren’t independent of each other.
UPDATE:
A few more details of how I obtained the distributions of transmitted material. I started with a set of 1374 parent-offspring transmissions that we had information for.
For each transmission I took the observed set of crossover events for each chromosome. If a chromosome had no crossovers, with probability 1/2 the parent transmitted the entire grandparental chromosome, otherwise they transmitted nothing for this chromosome.
If a chromosome had one or more recombination events in its transmission from a parent, both grandparents will have a contribution. We then have to decide who contributed what material based on the locations of the recombination events. The crossovers define a set of intervals transmitted together, which alternate between which grandparental material is transmitted. So for each transmission with probability 1/2 I make the parent transmit the grandparental corresponding to the odd inter-recombination intervals, else they transmit the even inter-recombination intervals.
Thus my simulations represent real transmissions, the only simulated part is the realization of Mendelian transmission (i.e. the 50/50 transmission probabilities). This means that the chromosome specific plots are not really simulations, and truly reflect these transmission data (each transmission contributing two datapoints, corresponding to the two grandparents).
My whole genome simulations are simulations, that assume independence of mendelian transmission across chromosomes. Only strong selection on viability/meiotic drive at individual loci could violate this assumption, and in general their is little evidence for this in humans. Given this assumption I can simulate vast numbers of transmitted autosomes due to the different realizations of Mendelian segregation across chromosomes. These represent pseudo-samples, in the sense that they only reflect the variation in the placement of recombination events across our 1374 parent-offspring transmissions. But overall I think this is not a bad way to approximate the distribution of transmitted material. It won’t be quite right in the very extreme tails, and that would need data on vast more transmissions.


u turned me into a cohen 🙂 (khan, not kahn)
sorry, must have been an ecoptic gene conversion (corrected).
Andrea Wishart asked me on twitter the good question of why the different chromosomes aren’t independent. I answered her there, and append a link to our conversation here in case it is of interest to others: http://twitter.com/pickleswarlz/status/392100767571509248
How much real family data do we have available to see if reality matches your simulations? I know Razib has done his family, and I’ve done my daughter, and Cece Moore (Your Genetic Genealogist) has access to quite a few families, all based on 23andMe data. Has23andMe done any analysis?
Hi Rosie,
Sorry, to be clear: my simulations use real recombination transmissions, based on family data we have for 1500 transmissions. They only thing simulated here is the pairing across chromosomes of the transmitted material.
Tim Janzen has done autosomal DNA tests on about 200 of his relatives and maintains a detailed spreadsheet showing the amount of shared DNA for all the different relationships though he doesn’t have data on grandparents. The link to download the spreadsheet can be found in this section of the ISOGG WIki article on autosomal DNA statistics: http://www.isogg.org/wiki/Autosomal_DNA_statistics#Tim_Janzen.27s_statistics_categorized_by_genealogical_relationship
The downloadable spreadsheets on his website for close relatives do contain grandparent-grandchild results, but would not exclude the X, and not very many of them.
would be nice to see standard deviations, not just ranges
Thanks for the link. Those are interesting but don’t really give a sense of the distribution. I’ll post similar results for a variety of transmissions some time soon.
I’ll ask TIm Janzen if he can add the standard deviations to his spreadsheet. We will also see if we can add the distribution of the shared DNA for all the relationship levels to our Wiki page. I look forward to Graham’s new calculations. It would be interesting to see more real-life data based on known relationships from different populations.
just to be clear, for the last two histograms, should i read it as fraction divided by 2 when comparing any focal grandparent to their grandchildren? e.g. can i rewrite your setence as: “Roughly 5/1000 grandchildren obtain 10% their autosome from one paternal grandparent” (expected value being 25%)
yep, that’s right (if my sims are correct).
thanks! incredibly long tail indeed.
“Doing this, we find that the probability that a male transmits every chromosome without recombination is 8.8 x 10^(-16), and this probability is substantially lower than that in females at 2.8×10^(-23).”
You mean higher here right?
Thanks, updated.
I have a question about this statement :
Roughly 5/1000 fathers transmit less than 20% of one paternal grandparent’s autosome to the next generation!.
So suppose a paternal grandmother contributed 18% of his genome to me. And suppose my paternal grandmother belongs to some exotic nationality, like a Yanomami or a Frenchman. In colloquial-geneaological terms I would be characterised as “one quarter Yanomami”. But if my Yanomani grandmother’s autosomnal contribution was 18%, would this “one quarter” have any meaning in genetics ? And would a genetic ancestry test discover that one of my grandmothers was a Yanomami ? Or would it just say I am 18% Yanomami ? ( I deliberately cited the paternal grandmother to exclude analyses of the Y-chromosome or the mtDNA. )
The genetic assignment tests (such as used by 23and me) try to estimate what fraction of your ancestry come from particular “source” populations. So if they are well callibrated they predict the 18%.
That 25% is what you expect, with any real transmission having some noise around that, but it doesn’t have any meaning in genetics given the observed value. A practical outcome of the lower sharing you suggest is that you would expect to resemble that grandparent slightly less in traits that had a genetic component.
Thanks. A follow-up question : in this particular hypothetical the 18% were contributed by the paternal grandmother, precluding the possibility that the Yanomami grandparent shows up through the Y-chromosome or the mtDNA. But would ancestry tests, or any other genetic testing, still be able to discover that one of the grandparents was Yanomami ? Or could they only infer that some of your ancestors were Yanomami ? I assume how recent was the 18% contribution would be discernible in the testing.
Pingback: How Many Ancestors Share Our DNA? | Genetic Inference
Pingback: How many genetic ancestors do I have? | gcbias
1.) Why does the male distribution have a dip at 50%?
2.) When you write “fraction of of grandparental genome transmitted via females”, is that
a.) “the fraction of maternal grandparental genome transmitted to a grandchild of either sex”
or
b.) “the fraction of grandparental genome transmitted to a female grandchild”
or
c.) “the fraction of maternal grandparental genome transmitted to a female grandchild”?
Hi Jay,
Thanks for your questions.
“Why does the male distribution have a dip at 50%? ”
-Given the large chunks transmitted to the next generation by males it is unlikely that a male will transmit very close to 50% of their (say) paternal autosomal genome to the next generation. So he might transmit 40% of the paternal autosomal genome and 60% of his maternal genome (or vise versa). This induces a bimodality in the distribution. It’s there in the female distribution as well, it’s just not very visible with the histrogram midpoints I used, as females are less variable in how much they transmit. You can also see this effect in the individual chromosome plots. I’m somewhat jetlagged writing this so hopefully it makes sense.
Graham
The graphs show the fraction of the entire (autosomal) genome from a particular grandparent that was transmitted to the next generation via a females. This means the fraction of your (say) maternal grandmother’s genome that your mother transmits to you (or equally the maternal grandfather). The sex of the grandparent doesn’t matter (nor the sex of the grandchild), the female refers to the fact that it is transmitted by the mother (I think that is a in your list above). What matters here (for the autosome) is the sex of the transmitting parent. Hope this clears that up, and sorry for any confusion.
Graam
Any chance of noting standard deviations on these (e.g., 95% and 99% probability limits)? I’ve been discussing this with someone stuck on the theoretical limits 0-50% from a given grandparent as being a reasonable way to describe it (i.e., average of 25% but ranging from 0 to 50%… argh!). I suppose I should be able to derive it from the info above, but I’m afraid I’m not up to that level ;-)… Thanks for the posts, and I wish you’d undertake a few more along similar lines!
Not being a mathematician or geneticist, I’m ill equipped to fully understand your study. My simple question is whether it is possible for my father’s father’s mother (my great grandmother) to be 50% Native American when both myself and my father show no NA origins in our Ancestry DNA reports? Also, are 23andMe tests more or less accurate than Ancestry DNA?
Wow, excellent. This post has some frequency data that seems to be almost impossible to find elsewhere.
The histograms in figure 2 are hard to follow, since I don’t know what units are used for “amount transmitted”. I would naively expect the “amount transmitted” value to range from 0 (none) to 1 (all), where the 0 and 1 values mean no crossover, while values in between indicate the position of the crossover. The units you use are what? Number of base pairs?
From your data, “the probability that a male transmits every chromosome without recombination is 8.8 x 10^(-16)”, I assume I can take the 23rd root of this number, and calculate that the average probability of a chromosome being transmitted with no crossover is 22%, and thus the crossover probability of a single chromosome, on the average is 88% (for a male, and 89.5% in a female)
Glad you like the post. The units on “amount transmitted” are indeed the number of DNA base pairs. I thought it was interesting to leave it as this, as different chromosomes have very different sizes (eg. 1 vs 21), but I should have labelled it more clearly.
So your calculation is along the right lines but I think it isn’t quite right. This is because the 8.8 x 10^(-16) include the probability of failing to recombine AND transmitting a particular grandparental chromosome (a factor of 1/2 for each chromosome). So removing this, the probability of failing to recombine is 8.8 x 10^(-16)/(0.5^22)= 3.698 x10^(-9). Taking the 22 root of this (22 autosomes) gives 42% as the (geometric) mean probability of no recombination. The data underlying this is on Page 17 of this supplement (link: http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1000658#pgen.1000658.s001). You can divide the zero count by the sum of each row.
Thanks for you comment!
What are the chances of autosomal DNA from a grandparent not being transferred to any grandchildren? The grandparent is related to the parent but they don’t appear in 23&me, etc. matches.
very interesting! can we turn the question around and ask: From how many siblings would we need to have DNA sequence to fully reconstruct the complete sequence of their parents and grandparents based on overlaps among the inherited DNA chunks?
Hi, would you expect two children of the same parent to have the same fraction passed down (the recombinations are in the parent?)? Or are they independent samplings of the distribution above? (I’m trying to figure out the amount a parent has in common with someone, given how much her kids do.) Sorry if this is obvious! Thanks!
kids are independent samples of the distribution above. The recombination events contained in the the gametes, the sperm and eggs that formed each child, are independent.
Thank you very much!
Hi! If I understand correctly, then, say kid1 and kid2 have overlaps with a person (related to one grandparent and their mom) of amounts kid1 and kid2. Can one say that the probability of overlap of their mom with that person is
P(mom) ~ P_m(kid1/mom)P_m(kid2/mom)/mom^4 where i read off P_m from your plot(“probability of grandparental genome transmitted via females?”) . and then i just normalize? if one doesn’t have the cM overlap of the mom, just of the kids? Is this a way to take into account that two kids give more info than one? It would be great to hear if this interpretation is right or wrong. Thanks!!
Why do you have “2 times” in the expression 2 x 0.5^22=4.7×10^(-7)? I cannot find Mike Eisen’s twitter discussion. Would you mind explaining?
Pingback: Anzahl genetische Vorfahren pro Vorfahrengeneration – geteilte DNA der Ahnenreihen – PopGen.at
This was a phenomenal explanation. Thank you for sharing!
Pingback: New and Improved Autosomal Genetic Model | DNA Science