Our paper on a general method to detect divergent selection among populations on quantitative traits was published today in PLOS Genetics (Berg and Coop 2014, code, the paper was previously up on the bioRxiv and Haldane’s sieve). Our method uses the knowledge gained about the genetic basis of a trait from GWAS, which allows us to compare allele frequency patterns across many loci to determine whether there is evidence for that they are all responding in concert to some selective pressure.
The method is, in large part, an attempt to set up the correct null model for the distribution of among population variation, and to circumvent the problem of environmental confounding in trying to determine whether certain differences between human populations may have been caused by selection.
The central idea underlying our tests, as well as many quantitative genetics approaches that have been developed over the last 2 decades (see paper for references), is that we can use genetic marker data to get a good measurement of how much allele frequencies vary among populations. Then, if the environment can be held constant across populations, we ought to be able to use the allele frequency information to predict how large a range of phenotypic differences among populations we would expect under a neutral model of genetic drift. Once we have this prediction in hand, a neutrality test for that phenotype follows.
The difficulty in approaching this question for human populations is that we of course cannot hold the environment constant across human populations, and so we can’t tell from direct phenotypic measurements whether any unusually large differences among populations that are observed are due to a history of natural selection, genetic drift, or entirely due to environmental differences among populations.
To get around this problem, we rely on the GWAS data, which comes in the form of a list of a specific set of SNPs associated with a phenotype, as well an estimate of the additive effect size for each associated variant. Importantly, these alleles were identified as associated with the phenotype in a specific population. We use the estimated effect sizes, in combination with allele frequencies at these loci, to predict the mean value of the trait we would expect to observe in each population, under a model in which all effects are additive and the environment constant. We can then test whether these genetic values differ among populations more than we would expect under a model of genetic drift.
We think that this test is potentially a good way to look for divergent selection pressures acting on traits. We expect that over the coming decade approaches such as ours, and more sophisticated developments, will be applied to many different traits. This offers a fascinating chance to study the role of natural selection in shaping phenotypic diversity among populations, in a range of species including humans.
However, beyond saying that selection, and not merely genetic drift, have acted on the loci involved in the trait, the interpretation of a positive results is challenging. We spend some time in the paper discussing these difficulties. Many of these are not new problems and some of these issues relate more generally to why it is difficult to study adaptation in natural populations. That said, it seems worth reiterating these issues and caveats carefully, as it is easy, and potentially fraught, to over interpret differences among human populations.
To do so, we use our signal of selection involving human height as an example. One of our clearest signals of selection is on height GWAS loci between Northern and Southern Europeans, as previously reported by Turchin et al. Nature Genetics. Turchin et al. and our research shows that alleles involved in height, at least those identified to date, show greater, directional differences between northern and southern European populations than would be expected under a model of genetic drift alone.
The first thing to say is that this signal is really quite subtle, and even where selection has acted, drift and gene flow still undoubtedly contribute to the differences we observe. Moreover, most of these variants are polymorphic in most of these populations, so they say little about an individual, i.e., most the variance is still among individuals within populations rather than among populations.
Nonetheless, average observed height clearly differs among European populations in the direction suggested by Turchin et al. and our results based on genetic data. Therefore, it is plausible that the observed differences in height between European populations are partially genetic in nature and have been subject to selection. We do not know this for certain, however, because of the large environmental contribution, e.g., diet, to traits such as height (as well as most human traits). Certainly the large changes in height over the past centuries in response to changes in nutrition and health highlight the huge role of the environment in height (see graph below). It remains possible that the majority of the observed mean difference among populations is environmental. While the relative order of populations has remained relatively constant, some have changed their ordering substantially, suggesting that we have to be very cautious about interpreting between population differences.
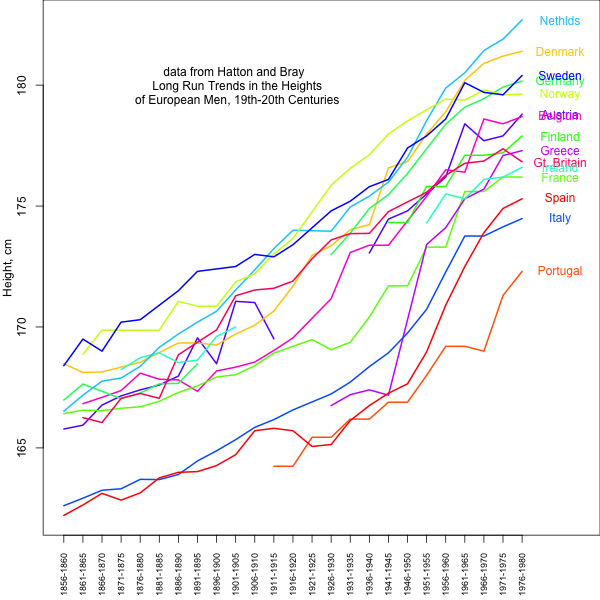
data from Hatton and Bray: Long Run Trends in the Heights of European Men, 19th-20th Centuries Econ Hum Biol. 2010 Dec;8(3):405-13. doi: 10.1016/j.ehb.2010.03.001. url: http://www.ncbi.nlm.nih.gov/pubmed/20399715 data extracted from pdf: http://privatewww.essex.ac.uk/~hatton/Tim_height_paper.pdf . Code here: https://github.com/cooplab/Height_over_time
Furthermore, only a small fraction of the variance in height within Europeans populations has been mapped to date (even less of the variance has been explained for many other traits). It is possible that as yet unmapped loci could change our understanding of the genetics of height in Europe. We could, for example find alleles that increase height which are common only in Southern Europe. While this is probably unlikely for height, it is exactly the case for skin pigmentation, where the partially convergent evolution of light skin pigmentation leads to confusing signals of adaptation that would be easy to misinterpret if we did not have a relatively good understanding of the genetics of skin pigmentation (see paper for details).
Also confusing the interpretation of the results is the potential for genotype by environment (GxE) or genotype by genotype interactions. Alleles may have different or even opposite effects in different environments and genetic backgrounds. While this cannot generate a false signal of selection, it does mean that the genetic values calculated for populations should not be treated as reliable phenotypic predictions. For height, it appears that many loci do seem to act in a reasonably additive manner and have consistent effects across populations. However, even given this trait, we have to be careful as we do not know if this holds for the subset of height GWAS loci that are driving our signal. Moreover, it is likely that traits other than height may have substantial GxE and so caution is warranted.
Thus, there is still a huge amount we do not understand about how selection and drift have shaped height, or any phenotype, within Europe. We also do not know that the differences in allele frequencies at variants associated with height reflect direct selection on height, or whether our observations are due to selection on some other phenotype (that may not even be on our radar in current environmental conditions, or may not differ among populations today). Even if selection acted directly on height, we would not know what the specific selection pressure that drove this difference. We also do not know the timing of this selection: whether it represents a long-term trend or is just a snapshot of some fluctuation, or whether selection took place in Europe or is the result of differential gene flow from populations who themselves diverged adaptively in height. While we expect that results from other fields, physical anthropology in particular, will be helpful, integrating these results to paint a more complete portrait of the evolution of human height will likely be challenging.
All of these caveats and concerns may seem overly cautious when expressed about studying the genetics of height differences among human populations. It seems quite likely that observed height differences among populations will be partially genetic in nature, and due in part to differential selection, consist with our and Turchin et al’s results. However, to establish this as a scientific finding, rather than a plausible hunch requires much more work. It is really quite humbling that we are only just beginning to understand the long-term role of selection and drift in shaping a phenotype as well studied as height. Undoubtedly, we will learn a lot over the coming decade about how drift, selection, and migration have shaped the genetic basis of phenotypes across populations, but these insights will only come about by the careful study of these phenotypes and the separation of genetic and environmental components.
While much of this probably seems obvious to most human geneticists, it is clear that there is huge public interest in genetic studies of human evolution and phenotypic differences and an almost equal potential for misunderstanding such studies. A sad case in point is the recent book by Nicholas Wade, which many reviews and blogs have already rightly criticized extensively. We hope that this blog post will help to layout some of the caveats that come with studying adaptation of complex traits even when good methods and GWAS are available.
Jeremy Berg and Graham Coop

I’ve had discussions with people recently about the steps you have to go through to convincingly demonstrate patterns of directional selection, and this is a clear and lucid description. Thanks!
Pingback: What we’re reading: Selection for heterozygosity in threatened seals, testing Fst outlier tests, and …? | The Molecular Ecologist
I’m not a scientist, just an interested amateur. But it seems to me that genetic inheritance can be completely hidden by nurture influence in a trait like height. Famine or inadequate food induced by family circumstances, climate catastrophe, politically-induced disaster, war or disease can result in entire generations, entire nations with stunted growth among peoples who would otherwise be taller. I’m 6′ tall, American of Irish ancestry who has recently found myself working with Chinese people taller than I am, taller than their own parents by well over 12 inches, despite the mostly-true stereotype of short Asians. In times of famine, circumstances can prevent a basic trait like height from being expressed. Being tall and large-physiqued could even be a temporary disadvantage until food supplies return to normal. (Speaking of the Irish, as I learn more about my own genealogy, I’m sure I’ll discover that that my pre-immigration ancestors were tiny, having been through a devastating famine of their own. Certainly the elderly relatives I’ve met in Ireland are all quite small, while the American generations are all much taller than even their Irish-American parents who invariably married other short Irish-Americans.)
Could a prolonged, multi-generational famine wipe out genes for tallness? Do countries which become better-fed always get taller?
Pingback: Polygenic scores and tea drinking | gcbias